Hongmei Qiao, Xiaoxuan Zhou, Yuchong Yi, Liufeng Wei, Xiuming Xu, Pengfei Jin, Wenyue Su, Yulin Weng, Dingtian Yu, Shanshan He, Meiping Fu, Chengcheng Hou, Xiaobao Pan, Wenqing Wang, Yuan-Ye Zhang, Ray Ming, Congting Ye*, Qingshun Quinn Li*, Yingjia Shen*
Current Biology
https://doi.org/10.1016/j.cub.2024.07.010
Published: 19 August 2024
Abstract
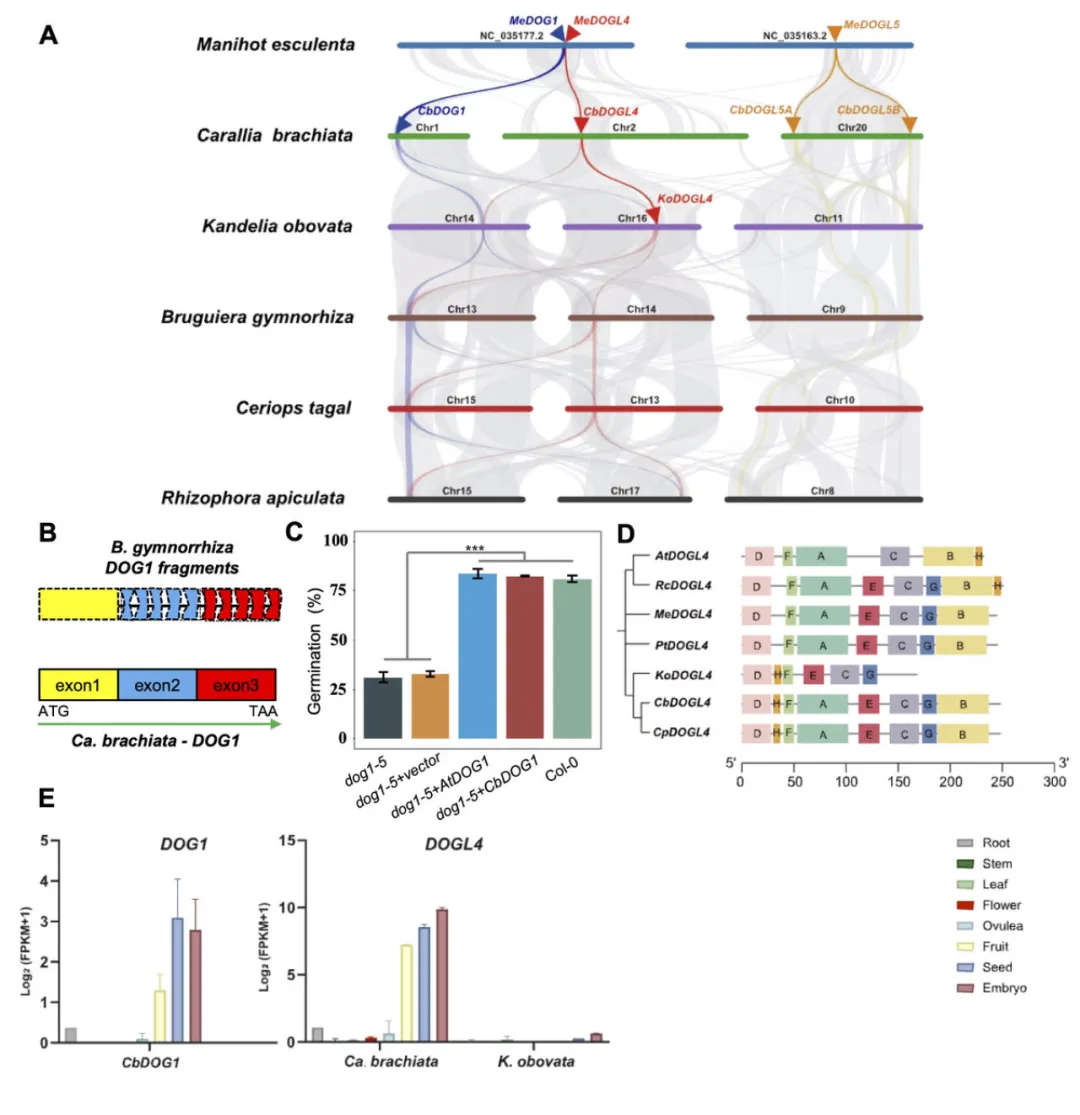
Vivipary is a prominent feature of mangroves, allowing seeds to complete germination while attached to the mother plant, and equips propagules to endure and flourish in challenging coastal intertidal wetlands. However, vivipary-associated genetic mechanisms remain largely elusive. Genomes of two viviparous mangrove species and a non-viviparous inland relative were sequenced and assembled at the chromosome level. Comparative genomic analyses between viviparous and non-viviparous genomes revealed that DELAY OF GERMINATION 1 ( DOG1 ) family genes (DFGs), DFGs ), the proteins from which are crucial for seed dormancy, germination, and reserve accumulation, are either lost or dysfunctional in the entire lineage of true viviparous mangroves but are present and functional in their inland, non-viviparous relatives. Transcriptome dynamics at key stages of vivipary further highlighted the roles of phytohormonal homeostasis, proteins stored in mature seeds, and proanthocyanidins in vivipary under conditions lacking DFGs. . Population genomic analyses elucidate dynamics of syntenic regions surrounding the missing DFGs. . Our findings demonstrated the genetic foundation of constitutive vivipary in Rhizophoraceae mangroves.